Research Projects
Metabolic interactions in spatially structured microbial communities
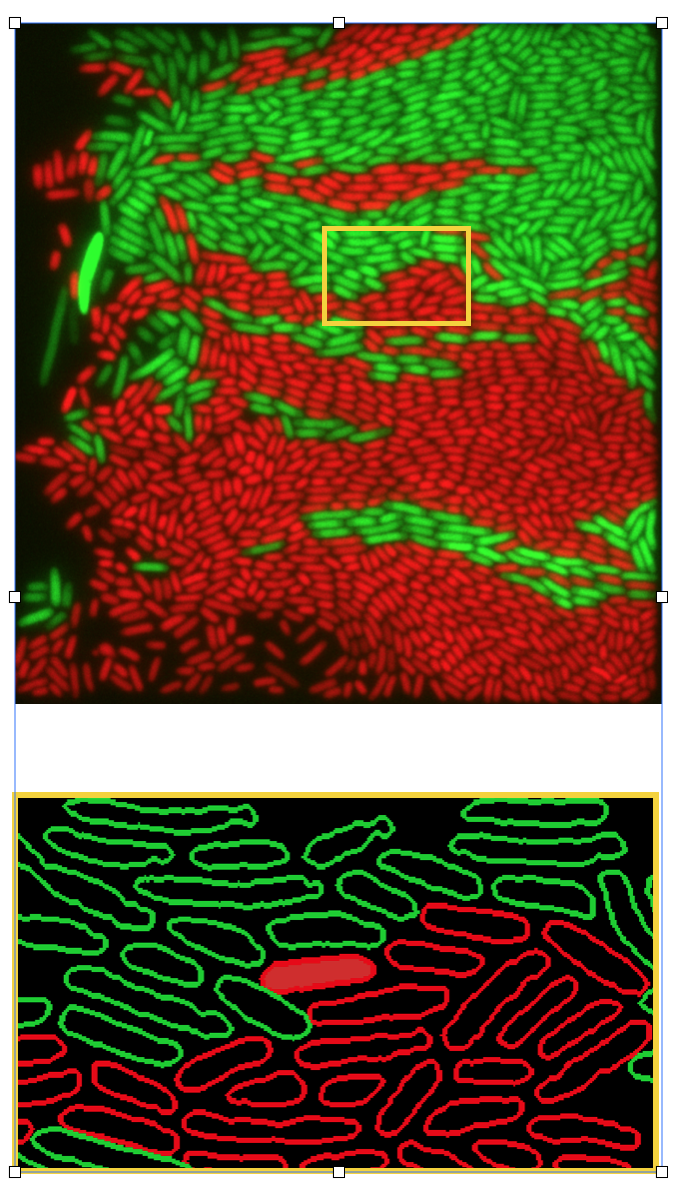
- Role of metabolic interactions in shaping the spatial distribution and growth dynamics of biopolymer (e.g., alginate and chitin) degrading populations in microbial communities (Glen, Michael, Astrid, external pageJulia Schwartzmancall_made, Johannes Keegstra (Roman Stocker group))
- Metabolic interactions with non-growing bacteria (Gatwa, Kim)
- Evolution of cell-cell interactions in spatially structured bacterial communities (Divvya and Emanuele)
- Effect of contact-dependent antagonism on nutrient flow in bacterial communities (Astrid and Glen)
- Effect of bacterial motility on the efficiency of metabolic interactions (Roy)
- Role of bacterial stalks in metabolic interactions (Roy)
- Antibiotic tolerance in microbial cross-feeding communities (Guga)
- Pattern formation in multi-component biofilms (Guga, and external pageDave Johnson groupcall_made)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
Group behavior and collective nutrient breakdown in microbial groups

- Impact of nutrient complexity on transitions between group and solitary behaviors in clonal groups (Glen, Johannes Keegstra (Roman Stocker group))
- Links between physiology, enzymatic capabilities and the strength of group behaviors (Glen and Ali Ebrahimi (external pageOtto Cordero groupcall_made))
- Molecular and regulatory underpinnings of bacterial behavior during stages of biopolymer degradation (Glen, Astrid, Michael, external pageJulia Schwartzmancall_made, Johannes Keegstra (Roman Stocker group))
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
Evolution of microbial interactions
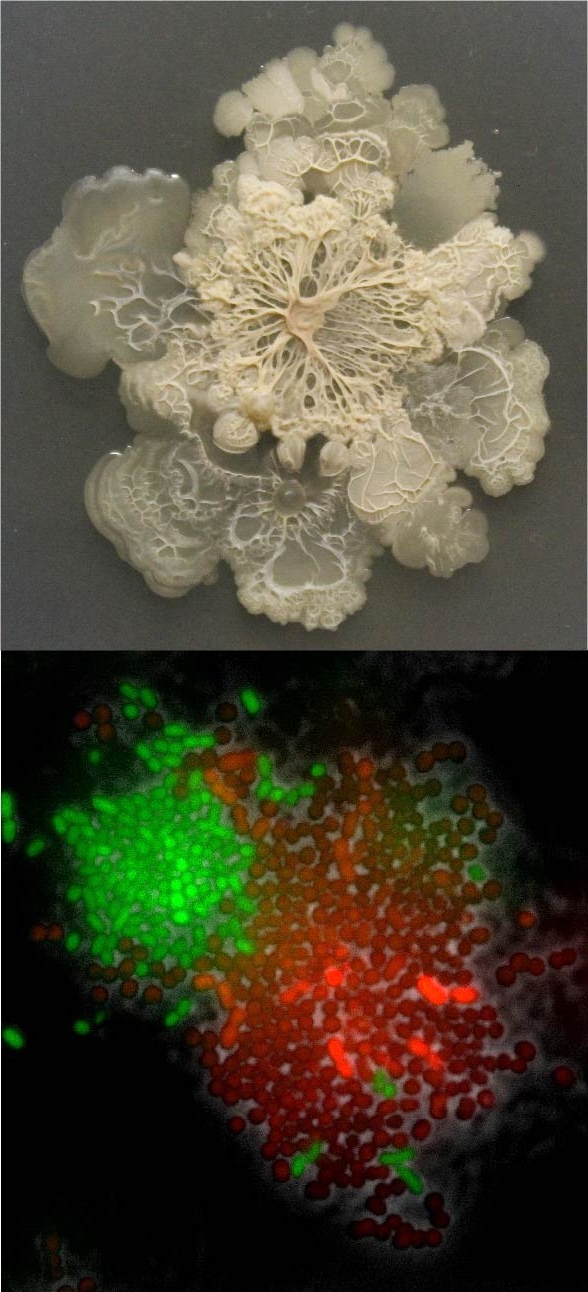
- Experimental evolution of aggregation increases intracellular synergy in microbial populations (Glen and Astrid)
- The evolution of cell-cell interactions in spatially structured synthetic microbial communities (Divvya and Emanuele)
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
- Factors that determine the degree of metabolic distribution in microbial ecosystems (Alyssa)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
Quantitative infection biology and antibiotic tolerance
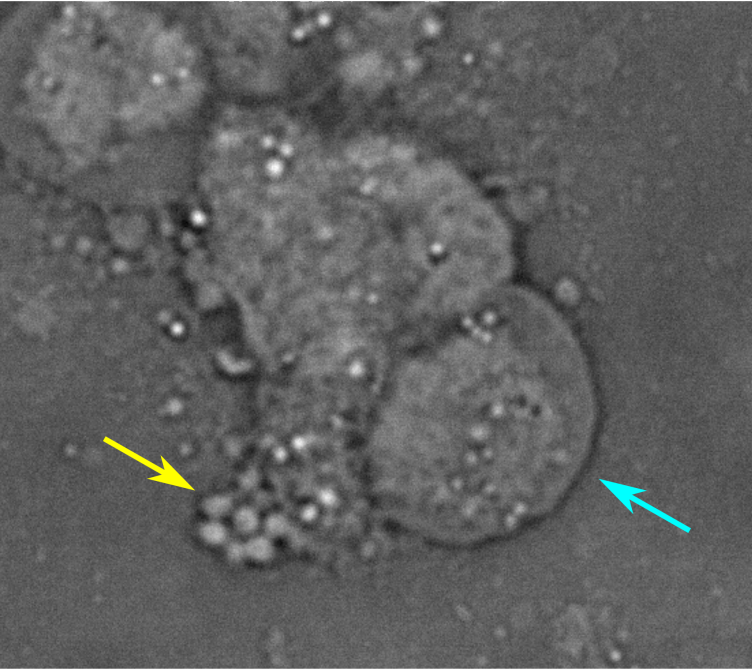
- Phenotypic heterogeneity in salmonella populations (Alyson and Divvya)
- Activities of salmonella sub-populations using single cell transcriptomes (Alyson and Astrid)
- Antibiotic tolerance in microbial cross-feeding communities (Guga)
Nutrient limitation and co-limitation
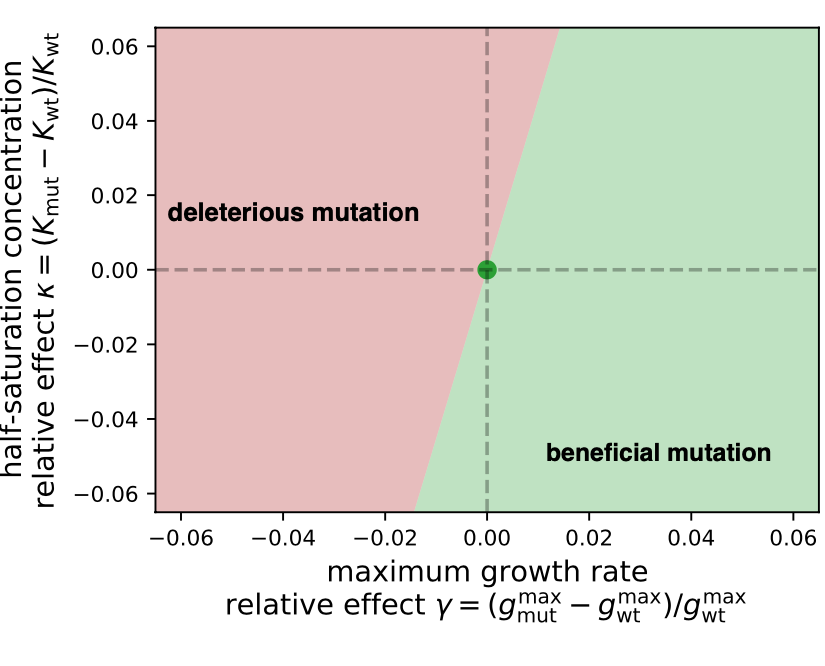
- Phenomenological descriptions and models of microbial growth in nutrient co-limitation (Noelle, external pageMichael Manhartcall_made, Justus)
- Quantitative competition dynamics and resource interaction in microbial communities (Justus, external pageMichael Manhartcall_made, Noelle)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
- Changes in nutrient-use efficiency as a function of a second limiting nutrient (Noelle, external pageMichael Manhartcall_made)
- Molecular underpinnings of nutrient limited microbial growth (Noelle, Juanita Lara Gutierrez (Roman Stocker group))
Principles of metabolic pathway distribution
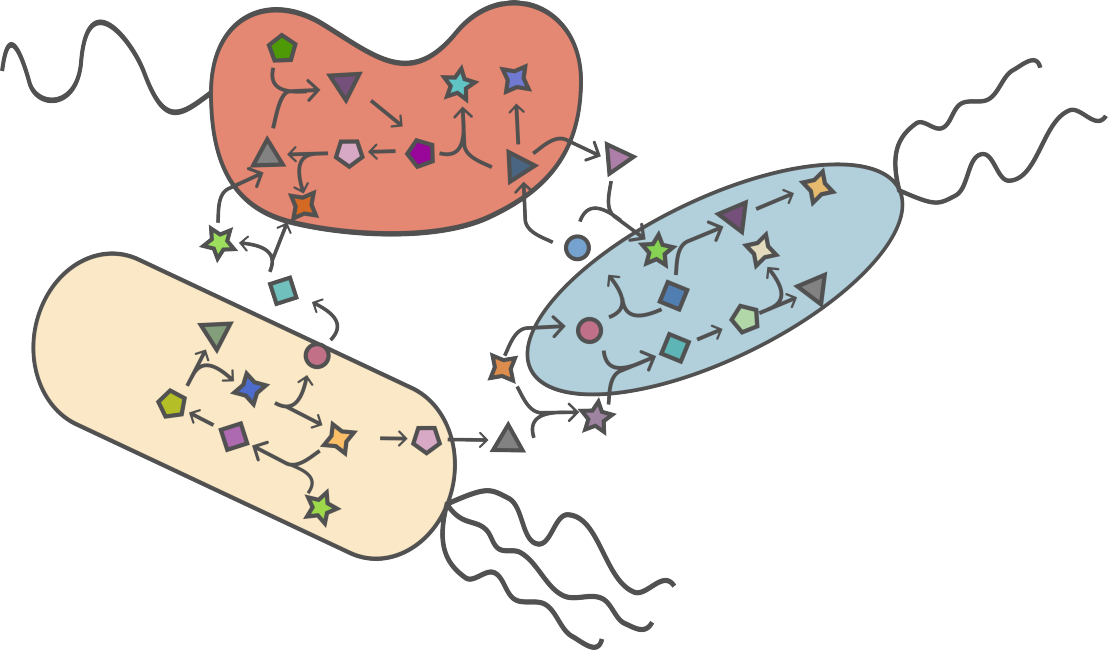
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
- Factors that determine the degree of metabolic distribution in microbial ecosystems (Alyssa)
Cell lysis as a strategy for nutrient acquisition
- Contact-dependent antagonism and its role in nutrient acquisition in microbial communities (Astrid and Glen)
- Mathematical modeling of lysing dynamics and the resulting nutrient flow (Astrid and François Peaudecerf (Roman Stocker group))
- Bacterial adaptation and global effects of contact-dependent antagonism (Astrid)
- Effect of phage lysis in cross-feeding communities (Emanuele)
Method development

- Analysis software for live-cell imaging data (Franziska (ETH SIS), Glen, Astrid, Olga)
- Microfluidics platform combining measurements of single cell growth and ecophysiology (Glen, Astrid, Fabai Wu external page(Victoria Orphan group)call_made)
- Measurement techniques for studying community properties and ecosystem functions (CO2) in microbial communities
- Biosensor for cyanotoxins (Oliver Brandenberg (external pageEawag Uchemcall_made), Elisabeth Janssen (external pageEawag Uchemcall_made))
- Techniques for measuring protein post-translational modifications in microbiome samples (Noelle, Ludovic Gillet, Paola Picotti group)
- Anaerobic microfluidics (Alyson, Markus Arnoldini (Emma Slack group), external pageSusan Schlegelcall_made (Uppsala University))
- Microfluidic systems for exploring diverse microbial interactions (Stefano)
