Research
The focus of our research group is on basic questions of the ecology and evolution of bacteria: on biological diversity at the level of individual bacterial cells, on interactions within and between species, and on how bacteria cope with ever-changing environments. The group often works at the level of single cells, and asks how this perspective provides insights that could not be obtained by population experiments.
Metabolic interactions in spatially structured microbial communities
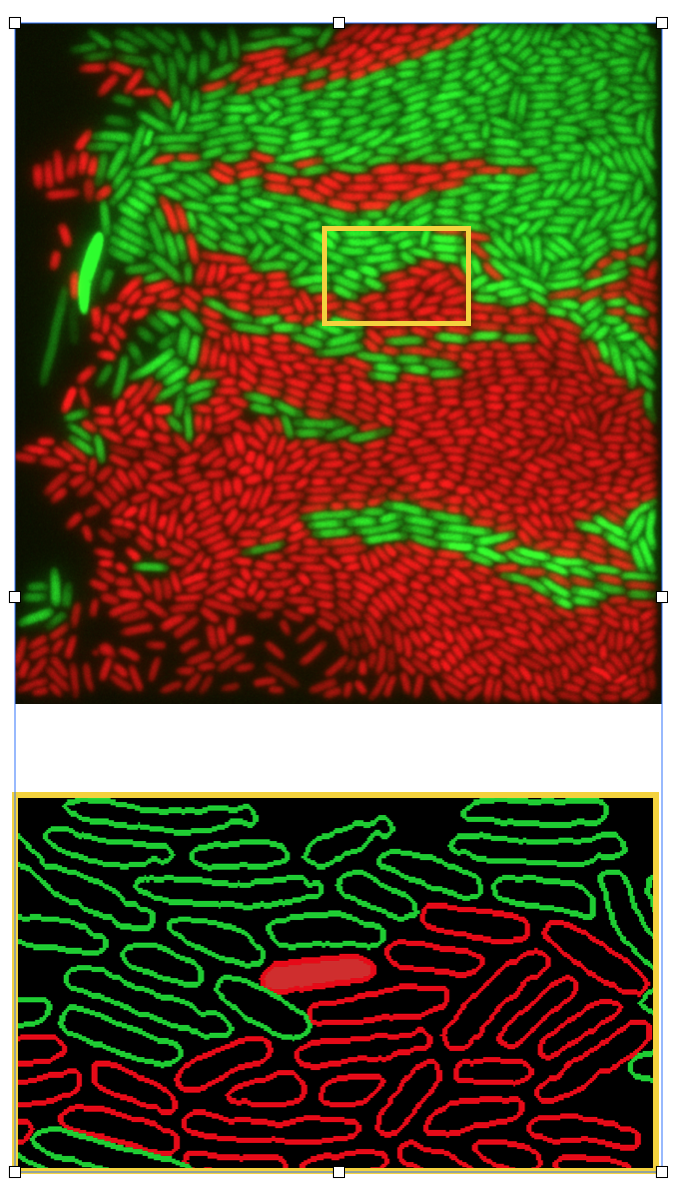
- Role of cross-feeding interactions in shaping the spatial distribution and growth dynamics of polymer degrading populations in microbial communities (external pagePriME collaborationcall_made: Glen, Michael, Astrid, Julia, Johannes Keegstra (Roman Stocker group), external pageJulia Schwartzmancall_made)
- Influence of metabolic cross-feeding on expression of polysaccharide degradation functions in marine microbial communities (external pageMichael Manhartcall_made, Noelle, Olga, and external pageSimon van Vlietcall_made)
- Cross-feeding interaction between L. plantarum & E. coli (Gatwa, Kim, and Olga)
- Evolution of cell-cell interactions in spatially structured synthetic microbial communities (Divvya and Emanuele)
- Effect of contact-dependent antagonism on the nutrient flow in communities (Astrid and Glen)
- Effect of motility on the efficiency of metabolic interactions (Roy)
- Understanding how the cell-cell connections mediated by bacterial stalk can benefit metabolic interactions (Roy)
- Antibiotic tolerance in microbial cross-feeding communities (Guga)
- Pattern formation in multi-component biofilms (Guga, and external pageDave Johnson groupcall_made)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
Group behavior and collective nutrient breakdown in microbial groups

- Impact of nutrient complexity on transitions between group and solitary behaviors in clonal groups (Glen, Vanessa, Johannes Keegstra (Roman Stocker group))
- Links between physiology, enzymatic capabilities and the strength of group behaviors (Glen and Ali Ebrahimi (external pageOtto Cordero groupcall_made))
- Molecular and regulatory underpinnings of bacterial behavior during stages of biopolymer degradation (Glen, Vanessa, Astrid, Michael, external pageJulia Schwartzmancall_made, Johannes Keegstra (Roman Stocker group))
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
Principles of metabolic pathway distribution
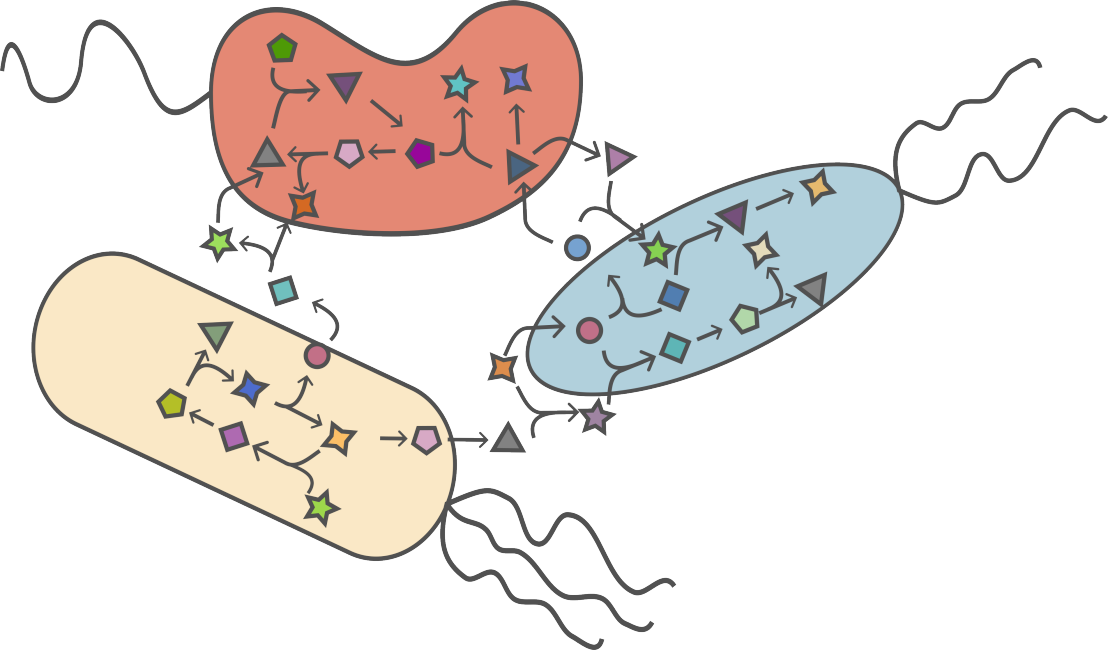
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
- Factors that determine the degree of metabolic distribution in microbial ecosystems (Alyssa)
Cell lysis as a strategy for nutrient acquisition
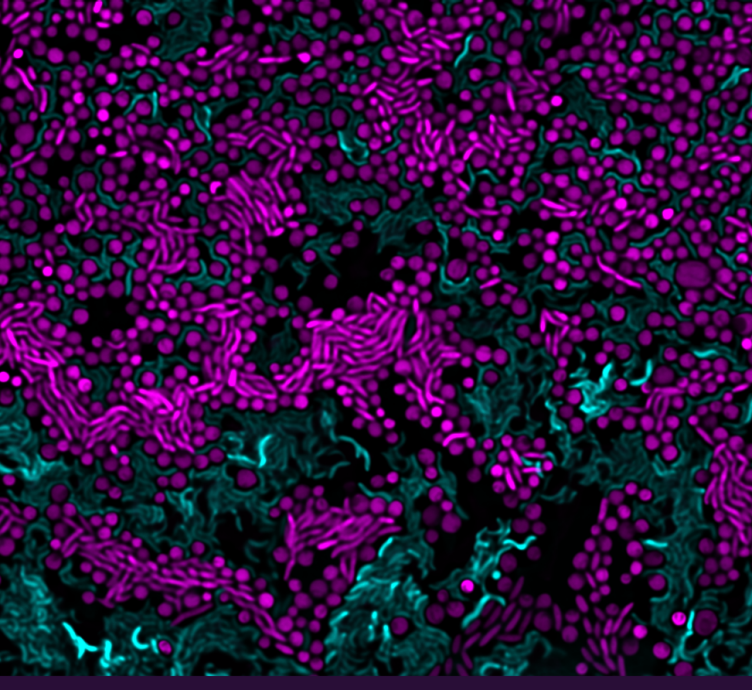
- Contact-dependent antagonism and its role in nutrient acquisition in microbial communities (Astrid and Glen)
- Mathematical modeling of lysing dynamics and the resulting nutrient flow (Astrid and François Peaudecerf (Roman Stocker group))
- Bacterial adaptation and global effects of contact-dependent antagonism (Astrid)
- Effect of phage lysis in cross-feeding communities (Emanuele)
Nutrient limitation and co-limitation
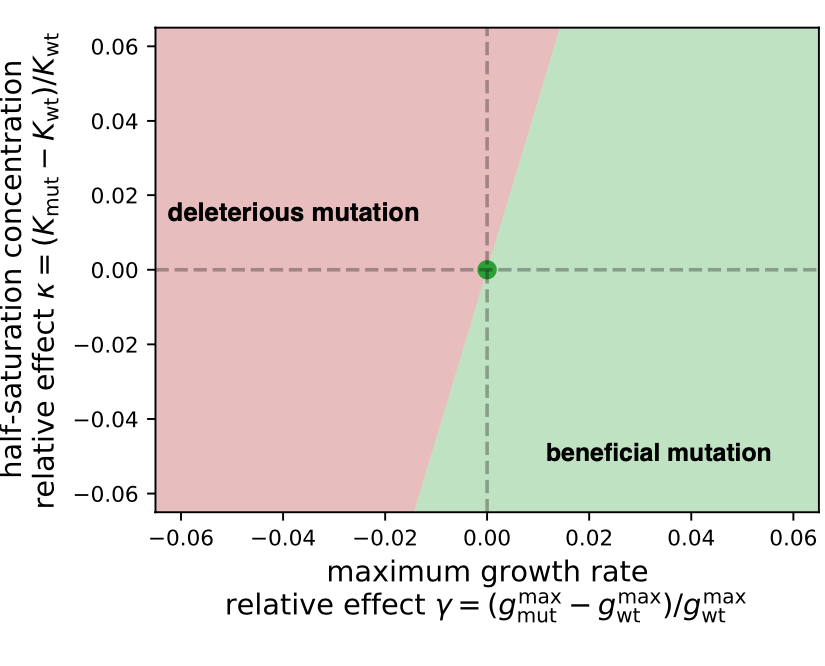
- Phenomenological descriptions and models of microbial growth in nutrient co-limitation (Noelle, external pageMichael Manhartcall_made, Justus)
- Quantitative competition dynamics and resource interaction in microbial communities (Justus, external pageMichael Manhartcall_made, Noelle)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
- Changes in nutrient-use efficiency as a function of a second limiting nutrient (Noelle, external pageMichael Manhartcall_made)
- Molecular underpinnings of nutrient limited microbial growth (Noelle, Juanita Lara Gutierrez (Roman Stocker group))
Evolution of microbial interactions
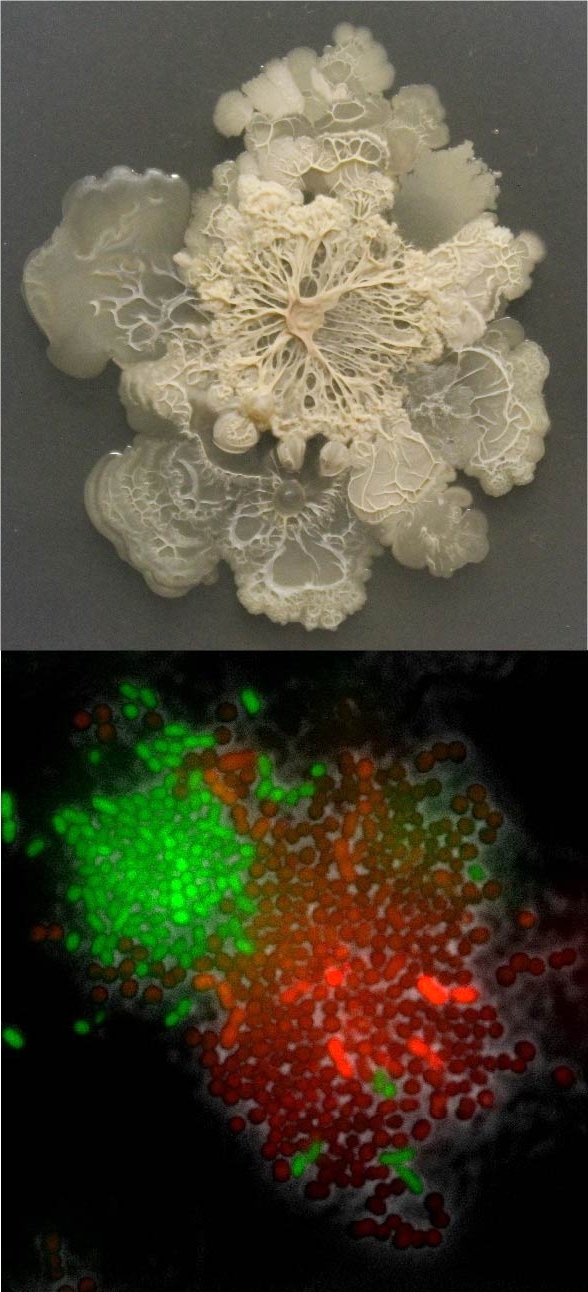
- Experimental evolution of aggregation increases intracellular synergy in microbial populations (Glen, Vanessa and Astrid)
- The evolution of cell-cell interactions in spatially structured synthetic microbial communities (Divvya and Emanuele)
- How proteome efficient metabolism of individuals underlies metabolic interactions and the collective metabolism of microbial communities (Matthias)
- Factors that determine the degree of metabolic distribution in microbial ecosystems (Alyssa)
- Evolution of microbial growth in nutrient limitation and co-limitation (Noelle, external pageMichael Manhartcall_made)
Quantitative infection biology and antibiotic tolerance
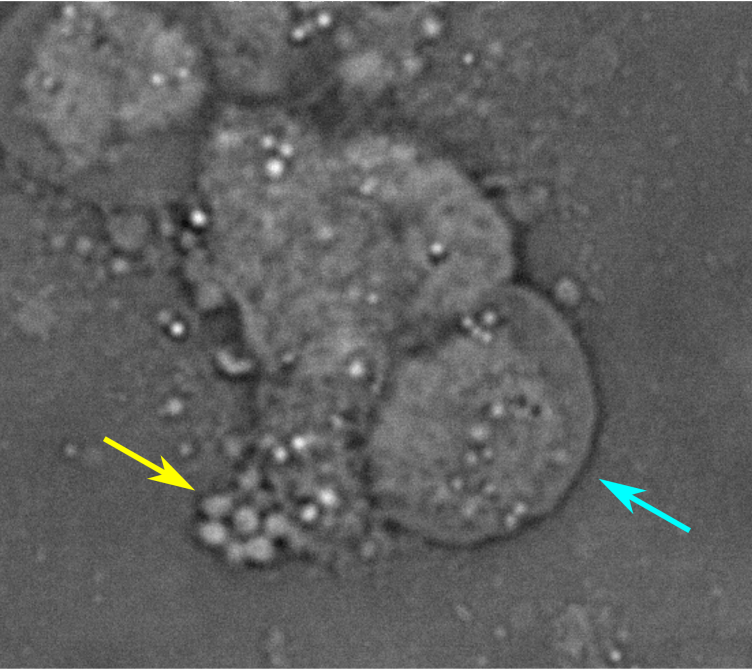
- Phenotypic heterogeneity in salmonella populations (Alyson and Divvya)
- Activities of salmonella sub-populations using single cell transcriptomes (Alyson and Astrid)
- Antibiotic tolerance in microbial cross-feeding communities (Guga)
Method development

- Analysis software for live-cell imaging data (Franziska (SIS), Janis (SIS), Glen, Astrid, Olga)
- Microfluidics platform combining measurements of single cell growth and ecophysiology (Glen, Vanessa, Astrid, Fabai Wu external page(Victoria Orphan group)call_made)
- Measurement techniques for studying community properties and ecosystem functions (CO2) in microbial communities
- Biosensor for cyanotoxins (Olga, external pageOliver Brandenbergcall_made (Eawag Uchem), external pageElisabeth Janssencall_made (Eawag Uchem))
- Techniques for measuring protein post-translational modifications in microbiome samples (Noelle, Ludovic Gillet, Paola Picotti group)
- Anaerobic microfluidics (Alyson, Markus Arnoldini (Emma Slack group), external pageSusan Schlegelcall_made (Uppsala University))
- Microfluidic systems for exploring diverse microbial interactions (Stefano)
